The Intricacies of Proteins and Their Design
The Wonders of Proteins
Proteins are composed of 20 simple amino acids through permutations and combinations. An amino acid sequence contains all the structural and activity information required to form a protein. An amino acid sequence can spontaneously fold into a unique three – dimensional structure and then perform specific functions within the cell. Some can bind to DNA to control gene switches, while others can recognize pathogens and initiate immune responses.
David Baker’s Pioneering Work
In 1993, David Baker of the University of Washington, one of the 2024 Nobel Prize in Chemistry laureates, began developing the Rosetta software to unravel the mystery of protein folding. At the same time, Baker also embarked on the challenge of “de novo protein design”. Compared to predicting protein structures, designing a protein from scratch requires scientists to deduce the DNA sequence from a protein with a specific shape. In 2003, David’s team designed the first protein that did not originally exist in nature, named Top7. Although this protein folded into the desired shape, it had no function.
Baker’s Current Role and Impact
Baker is now the director of the Institute for Protein Design at the University of Washington. He co – founded 21 companies, among which Xaira Therapeutics is the most renowned, having received over $1 billion in support to transform the research from his laboratory into drugs.
The Promise of Protein Design in Solving New Problems
Addressing New Challenges with Protein Design
On October 18, local time, in an interview with the biopharmaceutical industry media Endpoints, Baker talked about the importance of de novo protein design. “Proteins can perform an amazing range of functions, evolving over millions or billions of years to solve problems. Today, new problems have emerged. In the medical field, we are living longer, and thus new diseases have emerged. New pandemic viruses can emerge at any time. Outside of medicine, humans are warming the planet and creating pollution. The promise of protein design is to be able to design new proteins to solve current problems, as well as those related to the natural selection process of proteins in nature.”
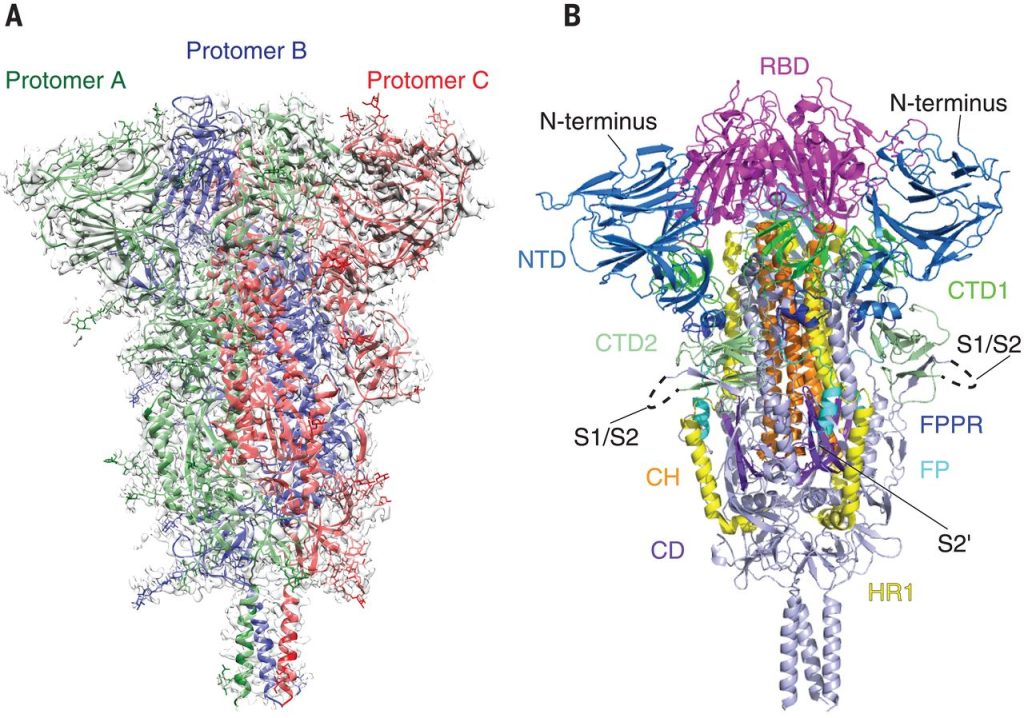
AI – designed Proteins Transforming Medicine and Technology
According to a report in Science on October 16, AI – designed proteins can transform medicine and technology. New tools have enabled researchers to produce designed proteins for vaccines and cancer treatments, artificial pollution – eliminating enzymes, and molecular components that can promote mineral growth. For example, shortly after the outbreak of COVID – 19 in 2020, researchers at the University of Washington designed proteins that attached to specific parts of the SARS – CoV – 2 spike protein and prevented the virus from penetrating human cells. Identifying this part of the spike protein enabled them to design a vaccine that arranged dozens of copies of the key protein part around a protein core to train the immune system to recognize and inactivate the same structure on SARS – CoV – 2. After successful human trials, this vaccine, named SKYCovione, was approved for use in South Korea and the UK last year, although its production has been put on hold due to the decline of the pandemic. Researchers at the University of Washington are working on other vaccines, including a broad – spectrum influenza vaccine that could eliminate the need for annual booster shots, and a vaccine against respiratory syncytial virus, a major killer of infants and the elderly.
The Outlook on AI – optimized Compounds
Baker’s Perspective on AI – optimized Compounds
When asked whether he was optimistic or skeptical about AI – optimized compound methods, Baker said, “If you want to predict whether a compound will pass clinical trials, you need the compound, hundreds of thousands of trials, and know exactly what happened in each trial to train a very effective model. We clearly don’t have this data.” He believes there are two paths of development. “The first is to identify substitutes that may achieve long – term success and then optimize them, such as targeting a certain amount of surface hydrophobicity as a structural substitute. The second is to generate relevant datasets. No entity on Earth can conduct 100,000 clinical trials and collect data. Large pharmaceutical companies have a lot of internal data on different compounds that failed in the drug development process. Interestingly, some companies are using this data for training, and the success will depend on the extensiveness of the dataset.”











































Discussion about this post